Predict fish phenology: advanced
Morgan Sparks, Bryan M. Maitland
Source:vignettes/Predict_phenology_advanced.Rmd
Predict_phenology_advanced.RmdOverview
The function predict_phenolgy() is purposefully kept
fairly simple so it can be expanded to iterate across datasets.
Typically, this will take the form of loops or vectorizing in R
(functions of the map() family from purrr
and apply() family from base). There are
many reasons why you might want to iterate, including predicting over
multiple years, multiple sites, predicting hatch or emergence
simultaneously, or even using multiple potential spawn dates. We’ll show
some examples below (moving from less to more complicated). First, we
load packages:
Multiple spawn dates
Given the example we used in the Predict Phenology-Basic vignette, perhaps we now want to expand it to predict multiple spawn dates (e.g., the early, peak, and late part of the spawning distribution) for the Woody Island population in Lake Iliamna. Using Table 1 from Sparks et al. (2019) we see that the spawning range is between August 14 and September 3rd, with the peak occurring August 18th. We can set these dates up as a vector to run the model over.
WI_spawn_dates <- c("1990-08-14", "1990-08-18", "1990-09-3")The we will go through the typical process of getting the model parameterized as we did previously.
sockeye_hatch_mod <- model_select(
author = "Beacham and Murray 1990",
species = "sockeye",
model_id = 2,
development_type = "hatch"
)Now we can automate the model. We’ll demonstrate two ways—a loop and a vectorized approach.
Looping
We will show an example of how to set up a for loop to
run through each element of the woody_island dataset.
# Loop storage objects
OUT_loop_all <- NULL
OUT_loop_d2h <- NULL
# Loop body
for (d in 1:length(WI_spawn_dates)) { # d will be our numerical iterator
# subset the element d of the vector and assign to object
WI_spawn <- WI_spawn_dates[d]
# predict phenology
WI_hatch <- predict_phenology(
data = woody_island,
dates = date,
temperature = temp_c,
spawn.date = WI_spawn,
model = sockeye_hatch_mod
)
### ALL output ###
# do this if we want to maintain all info predict_phenology
OUT_loop_all[[d]] <- WI_hatch
### A single element of output ###
# alternatively, subsets out output
temp <- tibble(matrix(data = NA, ncol = 2, nrow = 1)) # empty dataframe to add in data
colnames(temp) <- c("spawn_date", "days_2_hatch") # change column names
temp$spawn_date <- WI_spawn # assign spawn date
temp$days_2_hatch <- WI_hatch$days_to_develop # assign days to hatch
OUT_loop_d2h <- rbind(OUT_loop_d2h, temp) # row bind temp object and OUT object
}Inspect the loop output:
glimpse(OUT_loop_all)
#> List of 3
#> $ :List of 4
#> ..$ days_to_develop: int 71
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-08-14"
#> .. ..$ stop : Date[1:1], format: "1990-10-23"
#> ..$ ef_table : tibble [71 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>
#> $ :List of 4
#> ..$ days_to_develop: int 74
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-08-18"
#> .. ..$ stop : Date[1:1], format: "1990-10-30"
#> ..$ ef_table : tibble [74 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>
#> $ :List of 4
#> ..$ days_to_develop: int 110
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-09-03"
#> .. ..$ stop : Date[1:1], format: "1990-12-21"
#> ..$ ef_table : tibble [110 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>You can also see we made a tibble of dates and predicted days to hatch:
OUT_loop_d2h
#> # A tibble: 3 × 2
#> spawn_date days_2_hatch
#> <chr> <int>
#> 1 1990-08-14 71
#> 2 1990-08-18 74
#> 3 1990-09-3 110If you don’t want the full output of each run of
predict_phenolgy() and you just for example want the
predicted days to hatch, then it’s easier to access those elements
within the loop as demonstrated with the temp object
section demonstrated in the loop.
Vectorizing
Mapping
Vectorizing, while historically about prioritizing speed, is now more about making your code more readable (see Chapter 26 in R for Data Science and chapter 9 in Advanced R).
We’ll demonstrate using purrr::map(). If you are
unfamiliar with mapping but want to learn more about how it works and
it’s power, we highly recommend the linked chapters from R for Data
Science and Advanced R.
# map works by applying a function over a list (our vector is a very simple list)
# if you are familiar with apply() functions, map is essentially the same
results_map <- map(
WI_spawn_dates, # vector we are mapping over
predict_phenology, # function we are mapping with (note no "()"),
data = woody_island, # additional arguments required by predict_phenology
dates = date,
temperature = temp_c,
model = sockeye_hatch_mod
)
# we now have a list of lists the same as OUT_loop_all
glimpse(results_map)
#> List of 3
#> $ :List of 4
#> ..$ days_to_develop: int 71
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-08-14"
#> .. ..$ stop : Date[1:1], format: "1990-10-23"
#> ..$ ef_table : tibble [71 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>
#> $ :List of 4
#> ..$ days_to_develop: int 74
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-08-18"
#> .. ..$ stop : Date[1:1], format: "1990-10-30"
#> ..$ ef_table : tibble [74 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>
#> $ :List of 4
#> ..$ days_to_develop: int 110
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-09-03"
#> .. ..$ stop : Date[1:1], format: "1990-12-21"
#> ..$ ef_table : tibble [110 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>
# we can then access days to hatch easily as such
results_map |> map_dbl("days_to_develop")
#> [1] 71 74 110Apply
While tidyverse packages like purrr
have become quite popular, the base functionality of R has long had a
set of functions to do similar things as map. They are called the apply
family. Because hatchR is built using much of the
functionality of the tidyverse we opt for using
purrr:::map() over apply() functions, however
we will demonstrate a similar workflow as above using the apply
family.
# lapply() is the equivalent function as map() in the apply family
# they both output lists
results_lapply <- lapply(WI_spawn_dates, # vector we are mapping over
predict_phenology, # function we are mapping with (note no "()"),
data = woody_island, # additional arguments required by predict phenology
dates = date,
temperature = temp_c,
model = sockeye_hatch_mod
)You can see that lapply() and purrr::map()
output the same results:
glimpse(results_lapply)
#> List of 3
#> $ :List of 4
#> ..$ days_to_develop: int 71
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-08-14"
#> .. ..$ stop : Date[1:1], format: "1990-10-23"
#> ..$ ef_table : tibble [71 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>
#> $ :List of 4
#> ..$ days_to_develop: int 74
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-08-18"
#> .. ..$ stop : Date[1:1], format: "1990-10-30"
#> ..$ ef_table : tibble [74 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>
#> $ :List of 4
#> ..$ days_to_develop: int 110
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-09-03"
#> .. ..$ stop : Date[1:1], format: "1990-12-21"
#> ..$ ef_table : tibble [110 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>Iterating over multiple variables
An obvious extension of the example above is to use sites with multiple years of data and to predict hatch and emergence. In the example from the Predict Phenology-Basic vignette, we saw we had some days where the logger appeared to be out of the water, so let’s use a cleaner dataset.
In this example we will use temperature from the Crooked River, a
tributary of the Boise River in Idaho, USA, from a reach with known bull
trout spawning. The dataset is included as
crooked_river.
# look at data structure
glimpse(crooked_river)
#> Rows: 1,826
#> Columns: 2
#> $ date <dttm> 2010-12-01, 2010-12-02, 2010-12-03, 2010-12-04, 2010-12-05, 20…
#> $ temp_c <dbl> 1.1638020, 1.3442852, 1.2533443, 1.0068728, 1.2899153, 1.229158…
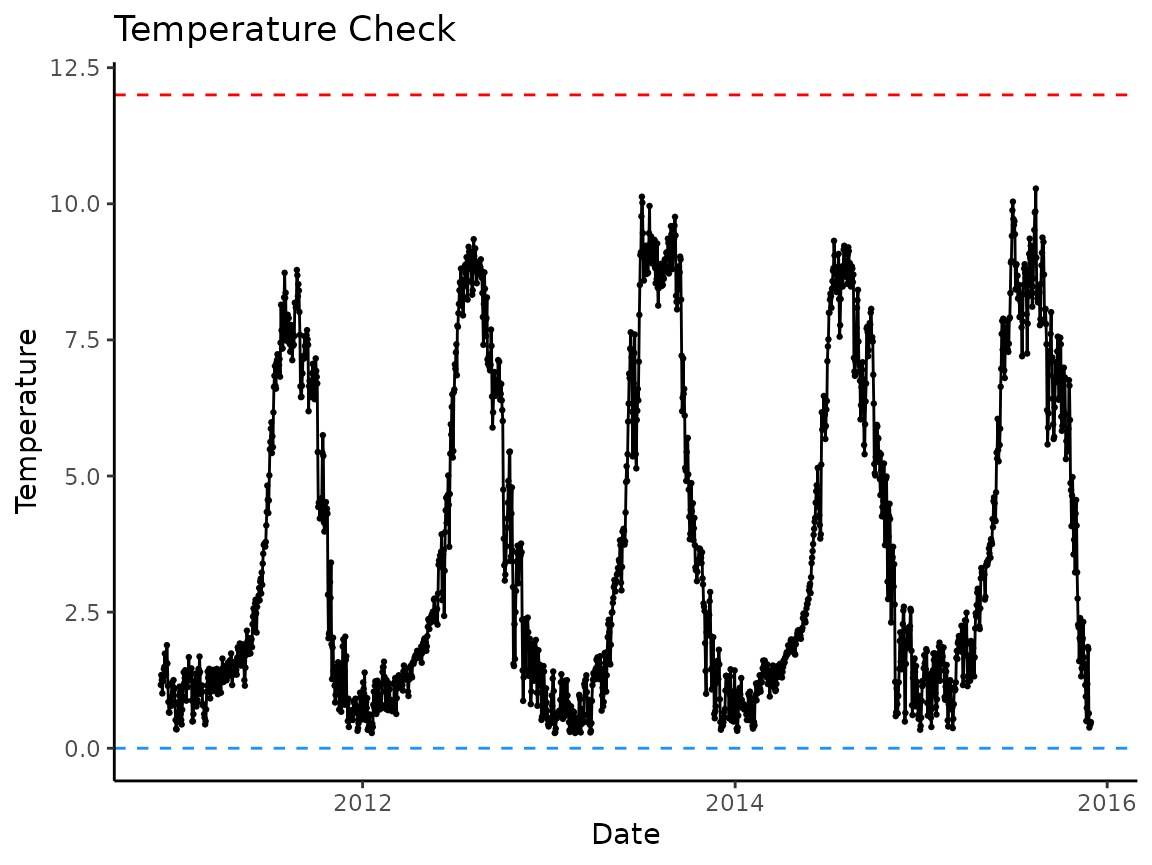
# visually check data
plot_check_temp(
data = crooked_river,
dates = date,
temperature = temp_c,
temp_min = 0,
temp_max = 12
)
We can use the functionality of dplyr to do a data check that shows the number of observations per year:
crooked_river |>
mutate(year = lubridate::year(date)) |>
group_by(year) |>
tally()
#> # A tibble: 6 × 2
#> year n
#> <dbl> <int>
#> 1 2010 31
#> 2 2011 365
#> 3 2012 366
#> 4 2013 365
#> 5 2014 365
#> # ℹ 1 more rowWe see we have 5 full years of data to use. For the sake of this exercise, let’s assume bull trout spawning in this system starts September 1st, peaks September 15th, and is mostly done by September 30th.
Remember, our data starts just barely in 2010, so our spawning years would be 2011-2015. However we may not have enough days of record to make predictions for 2015 so best to check that first.
First let’s just check to see if fish spawning in 2015 would have enough data to hatch and emerge.
# select bull trout hatch and emergence models
bull_hatch_mod <- model_select(
author = "Austin et al. 2019",
species = "bull trout",
model = "MM",
development_type = "hatch" # note we are using hatch model here
)
bull_emerge_mod <- model_select(
author = "Austin et al. 2019",
species = "bull trout",
model = "MM",
development_type = "emerge" # note we are using emerge model here
)
# set spawning dates in 2015
dates_2015 <- c("2015-09-01", "2015-09-15", "2015-09-30")
## hatch first
hatch_2015 <- dates_2015 |> purrr::map(
predict_phenology,
data = crooked_river,
dates = date,
temperature = temp_c,
model = bull_hatch_mod
)See how we receive 2 separate warning messages saying: “Fish did not accrue enough effective units to develop. Did your fish spawn too close to the end of your data? Spawn date is _____”. It tells us the spawn date we used so we can reference which date isn’t working if we use an string of multiple dates to predict over.
If fish aren’t hatching they definitely are not going to emerge, so we won’t use 2015.
Looping
We will build a nested loop to automate over model selection
variables and then over the dates. So the order of applying the
predict_phenology() function in the loop example below is
as follows:
- Predict hatch
- 2011 spawn dates
- …
- 2014 spawn dates
- Predict emergence
- 2011 spawn dates
- …
- 2014 spawn dates
## mini loop to make vector of dates
spawn_dates <- NULL
for (year in 2011:2014) {
temp_dates <- c(paste0(year, "-09-01"), paste0(year, "-09-15"), paste0(year, "-09-30"))
spawn_dates <- c(spawn_dates, temp_dates)
}
spawn_dates
#> [1] "2011-09-01" "2011-09-15" "2011-09-30" "2012-09-01" "2012-09-15"
#> [6] "2012-09-30" "2013-09-01" "2013-09-15" "2013-09-30" "2014-09-01"
#> [11] "2014-09-15" "2014-09-30"
### loop for prediction
OUT <- NULL
i <- 1 # build a counter for adding to our list of lists (OUT)
for (mod in c("hatch", "emerge")) { # loop for hatch and emerge
# here we just iterate model selection over the two hatch and emerge options
# in the first for loop
dev_mod <- model_select(
author = "Austin et al. 2019",
species = "bull trout",
model = "MM",
development_type = mod
)
for (d in 1:length(spawn_dates)) { # nested loop over spawn dates
# here we iterate over every date of spawn_dates but run this loop twice,
# once for each value of the first for loop
spawn <- spawn_dates[d] # get spawn date
temp <- predict_phenology(
data = crooked_river,
dates = date,
temperature = temp_c,
spawn.date = spawn,
model = dev_mod
) # notice we are calling the mod we set in the fist loop
OUT[[i]] <- temp
i <- i + 1 # add to your counter
}
}The output of this loop is again a list of lists. This time 24 lists
that comprise the elements of each predict_phenology()
iteration. Simply, the loop selects the hatch model, runs it for every
date in our vector (spawn_dates) and then switches over to
an emerge model and does the same thing. The point of having an iterator
(i) is that we can make a new list addition to
OUT for every combination of the dev_mod
(hatch and emerge) and the 12 dates we want to run—resulting in 24 total
individual lists that are appended to our OUT list of lists
as we walk through each iteration of the for loop.
If that sounds a bit complicated, it’s because it is. And this is where the vectorized approaches really shine.
A quick note about accessing lists. Even if you want to write loops instead of vectorizing, we do recommend accessing those results via a vectorized approach. It’s hard to pull out a lot of information from very large nested loops. You would probably write a custom loop to do so. However, those results are easily accessed by map, like so:
mapped_results <- OUT |> purrr:::map_df("ef_table")
mapped_results
#> # A tibble: 3,621 × 5
#> index dates temperature ef_vals ef_cumsum
#> <dbl> <dttm> <dbl> <dbl> <dbl>
#> 1 275 2011-09-01 00:00:00 6.65 0.0148 0.0148
#> 2 276 2011-09-02 00:00:00 6.45 0.0144 0.0292
#> 3 277 2011-09-03 00:00:00 6.46 0.0144 0.0436
#> 4 278 2011-09-04 00:00:00 6.66 0.0148 0.0584
#> 5 279 2011-09-05 00:00:00 6.88 0.0152 0.0736
#> # ℹ 3,616 more rowsThe above code outputs the ef_table element from every
one of your lists from each combination of spawn date and phenology type
into a dataframe like object which can then easily be plotted. Notice,
we now use purrr::map_df() because we know the list element
we are accessing is in the format of a dataframe type object (a
tibble).
Vectorizing
We’ll use the purrr function purrr::pmap() to vectorize
our multiple-input map function. Let’s again set up our iterative
variables. We’ll do the same workflow as the past example but now
exclusively with mapping.
# slightly cleaner spawn dates with map output is a list of vectors
spawn_dates <- map(
c(2011:2014), # year vector to map for custom function
function(year) { # custom function to make dates
c(
paste0(year, "-09-01"),
paste0(year, "-09-15"),
paste0(year, "-09-30")
)
}
) |>
unlist()
dev_mods <- map(
c("hatch", "emerge"),
model_select,
author = "Austin et al. 2019",
species = "bull trout",
model = "MM"
)
# we then set up a variable grid for all combinations of our models and dates
# it is very important to make the names of the columns in var_grid to match the
# arguments of the predict_phenology function (e.i., model = and spawn.date =)
var_grid <- expand_grid(model = dev_mods, spawn.date = spawn_dates)
head(var_grid)
#> # A tibble: 6 × 2
#> model spawn.date
#> <list> <chr>
#> 1 <spc_tbl_ [1 × 5]> 2011-09-01
#> 2 <spc_tbl_ [1 × 5]> 2011-09-15
#> 3 <spc_tbl_ [1 × 5]> 2011-09-30
#> 4 <spc_tbl_ [1 × 5]> 2012-09-01
#> 5 <spc_tbl_ [1 × 5]> 2012-09-15
#> # ℹ 1 more row
### multiple input mapping
crooked_predictions <- pmap(var_grid, # combos of variables to iterate over
predict_phenology, # function
data = crooked_river, # additional arguments required by function
dates = date,
temperature = temp_c
)Now that we’ve demonstrated both loops and mapping to get at iterating over multiple variables, let’s verify our results match.
# loop predictions
preds_loop <- OUT |> map_dbl("days_to_develop")
preds_pmap <- crooked_predictions |> map_dbl("days_to_develop")
# everything matches!
preds_loop == preds_pmap
#> [1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
#> [16] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUEGetting usable output
Because the basic output of these iterative processes are big lists
of lists, it can be a little annoying to access all that data. However,
this is again where the different flavors of map can be our friend.
We’ve already shown examples of how to access the predictions, but
predict_phenology() outputs much more data than just point
estimates.
First we’ll demonstrate an example of how we can take those prediction and use them to create an object that’s useful for plotting.
# get predictions
days <- crooked_predictions |>
map_dbl("days_to_develop")
# make a vector of what type of phenology we were predicting
# remember we ran hatch over 15 spawn dates then emerge over those same 15
phenology <- c(rep("hatch", 12), rep("emerge", 12))
# make a vector of our spawn dates replicated twice and turn into a timepoint
spawning <- rep(spawn_dates, 2) |>
ymd(tz = "UTC") # we do this because the crooked_river dataset is ymd_hms
# put them all together in an object
bull_trout_phenology <- tibble(phenology, spawning, days)
head(bull_trout_phenology)
#> # A tibble: 6 × 3
#> phenology spawning days
#> <chr> <dttm> <dbl>
#> 1 hatch 2011-09-01 00:00:00 88
#> 2 hatch 2011-09-15 00:00:00 106
#> 3 hatch 2011-09-30 00:00:00 124
#> 4 hatch 2012-09-01 00:00:00 89
#> 5 hatch 2012-09-15 00:00:00 104
#> # ℹ 1 more rowWe could then make a plot with our original data:
# filter crooked_river to correct size
crooked_river_11_15 <- crooked_river |> filter(date < ymd_hms("2015-06-01 00:00:00"))
ggplot(data = crooked_river_11_15, aes(x = date, y = temp_c)) +
geom_point(size = 0.25) +
geom_line() +
geom_point(
data = bull_trout_phenology,
aes(x = spawning, y = 10), color = "darkblue", shape = 25, size = 2.5
) +
geom_point(
data = bull_trout_phenology |> filter(phenology == "hatch"),
aes(x = spawning + days(days), y = 0),
color = "darkgreen", shape = 24, size = 2.5
) +
geom_point(
data = bull_trout_phenology |> filter(phenology == "emerge"),
aes(x = spawning + days(days), y = 0),
color = "darkorange", shape = 24, size = 2.5
) +
labs(
title = "Crooked River Bull Trout Developmental Phenology",
subtitle = "Blue = Spawn, Green = Hatch, Orange = Emerge",
x = "Date",
y = "Temperature"
) +
theme_classic()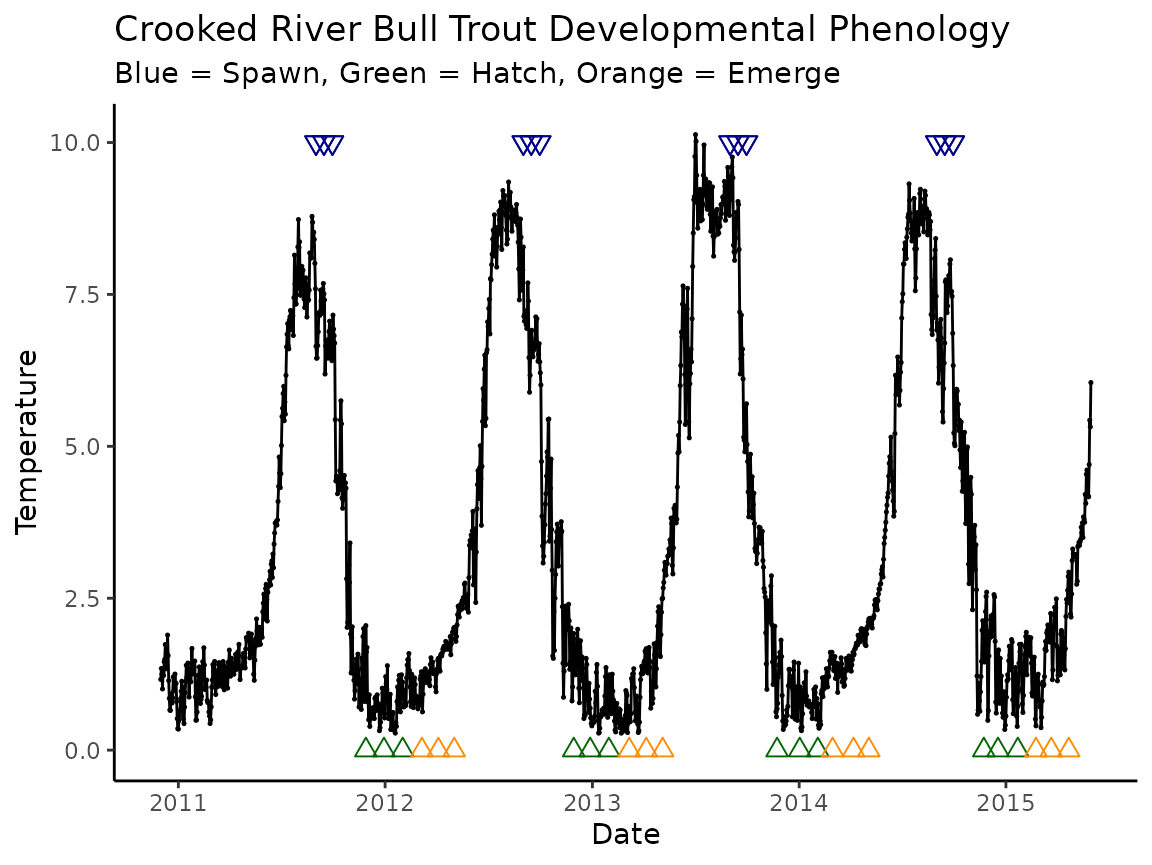
Not the best looking plot, but a good example of how to translate output to a visual representation. For better looking plots, please see the Advanced plotting vignette.
Other data types you might want might include the tibble with dates, temperature, and values and cumulative sums of effective values or the, alternatively, the developmental period.
# the tibble with all temperature and effective values for each phenological period
all_data <- crooked_predictions |>
map_df("ef_table")
head(all_data)
#> # A tibble: 6 × 5
#> index dates temperature ef_vals ef_cumsum
#> <dbl> <dttm> <dbl> <dbl> <dbl>
#> 1 275 2011-09-01 00:00:00 6.65 0.0148 0.0148
#> 2 276 2011-09-02 00:00:00 6.45 0.0144 0.0292
#> 3 277 2011-09-03 00:00:00 6.46 0.0144 0.0436
#> 4 278 2011-09-04 00:00:00 6.66 0.0148 0.0584
#> 5 279 2011-09-05 00:00:00 6.88 0.0152 0.0736
#> # ℹ 1 more row
# the phenological durations for each prediction set
development_period <- crooked_predictions |>
map_df("dev_period")
head(development_period)
#> start stop
#> 1 2011-09-01 2011-11-27
#> 2 2011-09-15 2011-12-29
#> 3 2011-09-30 2012-01-31
#> 4 2012-09-01 2012-11-28
#> 5 2012-09-15 2012-12-27
#> 6 2012-09-30 2013-01-29Naming your lists
A problem with this approach is that you end up with very large lists
of lists without labels which require you to remember the data structure
you feed to your mapping function. However, purrr does have a method to
get around this which is the purrr::set_names() function
which you would pipe into your mapping function.
# to use the simple Iliamna Lake example within a single season
WI_named_list <- WI_spawn_dates |>
set_names() |>
map(predict_phenology, # note we leave out the input row (WI_spawn_dates) because we are piping it in as input
data = woody_island, # additional arguments required by predict phenology
dates = date,
temperature = temp_c,
model = sockeye_hatch_mod
)
# you can now see each list is named according to its respective spawn date
glimpse(WI_named_list)
#> List of 3
#> $ 1990-08-14:List of 4
#> ..$ days_to_develop: int 71
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-08-14"
#> .. ..$ stop : Date[1:1], format: "1990-10-23"
#> ..$ ef_table : tibble [71 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>
#> $ 1990-08-18:List of 4
#> ..$ days_to_develop: int 74
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-08-18"
#> .. ..$ stop : Date[1:1], format: "1990-10-30"
#> ..$ ef_table : tibble [74 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>
#> $ 1990-09-3 :List of 4
#> ..$ days_to_develop: int 110
#> ..$ dev_period :'data.frame': 1 obs. of 2 variables:
#> .. ..$ start: Date[1:1], format: "1990-09-03"
#> .. ..$ stop : Date[1:1], format: "1990-12-21"
#> ..$ ef_table : tibble [110 × 5] (S3: tbl_df/tbl/data.frame)
#> ..$ model_specs : spc_tbl_ [1 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
#> .. ..- attr(*, "spec")=List of 3
#> .. .. ..- attr(*, "class")= chr "col_spec"
#> .. ..- attr(*, "problems")=<externalptr>
# therefore a single element could be accessed with its name and the $ operator
# if we wanted the dev_period for fish spawning on August 18th we would do the following
WI_named_list$`1990-08-18`$dev_period
#> start stop
#> 1 1990-08-18 1990-10-30This approach works well for the example above, but not so well with
our examples with purrr::pmap() because the variable grid
we create has a bunch of expressions which don’t work for naming lists.
However, if we were to do a purrr::pmap() example where it
was a site x dates combination, that still work and would name the lists
according to their site and then the date used to predict phenology
(e.g. named_list$site1$`date`$dev_period)