Overview
hatchR is an R package that allows users to predict hatch and emergence timing for wild fishes, as well as additional tools to aid in those analyses. hatchR is intended to bridge the analytic gap of taking statistical models developed in hatchery settings (e.g., Beacham and Murray 1990) and applying them to real world temperature data sets using the effective value framework developed by Sparks et al. (2019). hatchR is also available as an interactive web application at https://elifelts.shinyapps.io/hatchR_demo/.
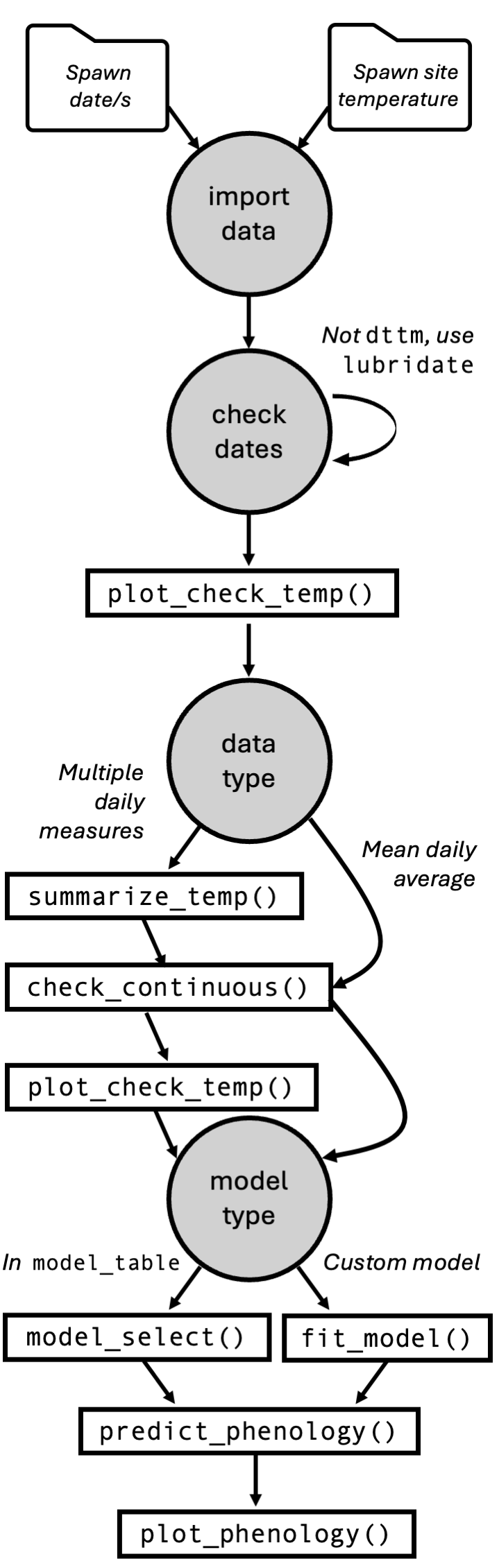
hatchR is designed to be flexible to achieve many applications. However, by virtue of being built as a scripting application, hatchR is able to tackle very large datasets relatively quickly and efficiently. That is, hatchR can be used to predict hatch and emergence timing for many fish species across many locations and years. These type of upstream analyses are covered in additional vignettes in this package. In this introductory vignette, we:
- describes input data requirements
- provide recommendations for importing data
- review basic data checks
- preview the full hatchR workflow covered in other vignettes
The generalized workflow for hatchR, including the steps below and in subsequent other vignettes, is shown below.
Input Data Requirements
hatchR requires three primary data inputs:
- Water temperature data
- Species-specific model parameters
- Spawning date or date ranges
Water Temperature Data
Water temperature datasets for wild fishes are often either (1) already summarized by day (i.e., mean daily temperature) or, (2) in a raw format from something like a HOBO TidbiT data logger where readings are taken multiple times per day, which can be summarized into a mean daily temperatures. Alternatively, daily water temperature predictions from novel statistical models (e.g., Siegel et al. 2023) could be similarly implemented.
Fundamentally, hatchR assumes you have input data with two columns:
- date: a vector of dates (and often times) of a temperature
measurement (must be class
DateorPOSIXct, which Tibbles print as <date> and <dttm>, respectively). - temperature: a vector of associated temperature measurement (in centigrade).
Additional columns are allowed, but these are required.
We expect your data to look something like this:
| date | temperature |
|---|---|
| 2000-01-01 | 2.51 |
| … | … |
| 2000-07-01 | 16.32 |
| … | … |
| 2000-12-31 | 3.13 |
hatchR assumes you’ve checked for missing records or errors in your data because it can work with gaps, so it’s important to go through the data checks discussed below, as well as your own validity checks.
hatchR can use values down to freezing (e.g., 0 °C), which returns extremely small effective values, and time to hatch or emerge may be > 1 year. In these cases, we suggest users consider how much of that data type is reasonable with their data. To learn more about this, see the section on negative temperatures in the Predict fish phenology: Basic vignette.
Dates and Times
Numeric temperature values are simple to work with in R, but dates and time can be tricky. Here we provide a brief overview of how to work with dates and times in R, but refer the user to Chapter 17 in R for Data Science (Wickham, Çetinkaya-Rundel, and Grolemund 2023) for a more in-depth discussion.
The lubridate package makes it easier to work with dates and times in R. It comes installed with hatchR, and can be loaded with:
There are three types of date/time data that refer to an instant in time:
- A date, which Tibbles print as <date>
- A time within a day, which Tibbles print as <time>
- A date-time is a date plus time, which Tibbles
print as <dttm>. Base R calls these
POSIXct, but that’s not a helpful name.
You can use lubridate::today() or
lubridate::now() to get the current date or date-time:
In the context of hatchR, the ways you are likely to create a date/time are:
- reading a file into R with
readr::read_csv() - from a string (e.g., if data was read into R with
read.csv()) - from individual components (year, month, day, hour, minute, second)
Reading in dates from a file
When reading in a CSV file with readr::read_csv(),
readr (which also comes installed with
hatchR) will automatically parse (recognize) dates and
date-times if they are in the form “YYYY-MM-DD” or “YYYY-MM-DD
HH:MM:SS”. These are ISO8601 date (<date>) and date-time
(<dttm>) formats, respectively. ISO8601 is an international
standard for writing dates where the components of a date are organized
from biggest to smallest separated by -. Below, we first load the
readr package and then read in a CSV file with dates in the
form “YYYY-MM-DD” and “YYYY-MM-DD HH:MM:SS”:
csv <- "
date,datetime
2022-01-02,2022-01-02 05:12
"
read_csv(csv)
#> # A tibble: 1 × 2
#> date datetime
#> <date> <dttm>
#> 1 2022-01-02 2022-01-02 05:12:00If your dates are in a different format, you’ll need to use
col_types plus col_date() or
col_datetime() along with a standard date-time format (see
Table
17.1 in R for Data Science (Wickham,
Çetinkaya-Rundel, and Grolemund 2023) for a list of all date
format options).
From strings
If you read in a CSV file using read.csv() from base R,
date columns will be formatted as a characters (<char>; e.g.,
"2000-09-01" or "2000-09-01 12:00:00"). You
will have convert to this column to a <date> or <dttm>.
lubridate helper functions attempt to automatically
determine the format once you specify the order of the component. To use
them, identify the order in which year, month, and day appear in your
dates, then arrange “y”, “m”, and “d” in the same order. That gives you
the name of the lubridate function that will parse your
date. For date-time, add an underscore and one or more of “h”, “m”, and
“s” to the name of the parsing function.
From individual components
Sometimes you have the components of a date in separate columns. You
can use make_date() or make_datetime() to
combine them into a date or date-time.
To show this, we’ll use the flights dataset that comes
with nycflights13, which is only installed alongside
hatchR when dependencies = TRUE in
install.packages(). We also make use of helper functions
from dplyr to select() and
mutate() columns.
flights |>
select(year, month, day) |>
mutate(date = make_date(year, month, day))
#> # A tibble: 336,776 × 4
#> year month day date
#> <int> <int> <int> <date>
#> 1 2013 1 1 2013-01-01
#> 2 2013 1 1 2013-01-01
#> 3 2013 1 1 2013-01-01
#> 4 2013 1 1 2013-01-01
#> 5 2013 1 1 2013-01-01
#> # ℹ 336,771 more rows
flights |>
select(year, month, day, hour, minute) |>
mutate(departure = make_datetime(year, month, day, hour, minute))
#> # A tibble: 336,776 × 6
#> year month day hour minute departure
#> <int> <int> <int> <dbl> <dbl> <dttm>
#> 1 2013 1 1 5 15 2013-01-01 05:15:00
#> 2 2013 1 1 5 29 2013-01-01 05:29:00
#> 3 2013 1 1 5 40 2013-01-01 05:40:00
#> 4 2013 1 1 5 45 2013-01-01 05:45:00
#> 5 2013 1 1 6 0 2013-01-01 06:00:00
#> # ℹ 336,771 more rowsTime Zones
Time zones are a complex topic, but we don’t need to delve too deep for our purposes. We will cover a few basic topics. For more information, see Chapter 17 in R for Data Science (Wickham, Çetinkaya-Rundel, and Grolemund 2023).
R uses the international standard IANA time zones. These use a consistent naming scheme {area}/{location}, typically in the form {continent}/{city} or {ocean}/{city}. Examples include “America/New_York”, “Europe/Paris”, and “Pacific/Auckland”.
You can find out what R thinks your current time zone is with
Sys.timezone():
Sys.timezone()
#> [1] "UTC"(If R doesn’t know, you’ll get an NA.)
In R, the time zone is an attribute of the date-time that only controls printing. For example, these three objects represent the same instant in time:
x1 <- ymd_hms("2024-06-01 12:00:00", tz = "America/New_York")
x1
#> [1] "2024-06-01 12:00:00 EDT"Unless otherwise specified, lubridate always uses UTC. UTC
(Coordinated Universal Time) is the standard time zone used by the
scientific community and is roughly equivalent to GMT (Greenwich Mean
Time). It does not have DST, which makes a convenient representation for
computation. Thus read_csv() converts to UTC, so if times
are shifted, then daily summaries can be incorrect. This only matters if
data are not daily yet, as.posixct() sets to local
time.
You can change the time zone in two ways:
- Change the time zone attribute with
force_tz() - Change the time zone for printing with
with_tz()
Importing water temperature data
Using readr::read_csv()
We recommend loading your data into R using
readr::read_csv(). We can load readr using:
In this and other vignettes, we will use two datasets that come
installed with the package: crooked_river and
woody_island. Each dataset is available to the user as R
objects once hatchR is installed and attached (see
?crooked_river or ?woody_island for more
information). The raw example data (.csv files) are stored in the
extdata/ directory installed alongside
hatchR. We may store the file paths to example
data:
path_cr <- system.file("extdata/crooked_river.csv", package = "hatchR")
path_wi <- system.file("extdata/woody_island.csv", package = "hatchR")After specifying path_* (using
system.file()), we load the example dataset into R:
We check the crooked_river dataset by running either
str() or tibble::glimpse() to see the
structure of the data. glimpse() is a little like
str() applied to a data frame, but it tries to show you as
much data as possible. We prefer tibble::glimpse() because
it is more compact and easier to read. tibble also
comes installed with hatchR.
Now we can check the structure of the crooked_river
dataset:
glimpse(crooked_river)
#> Rows: 1,826
#> Columns: 2
#> $ date <dttm> 2010-12-01, 2010-12-02, 2010-12-03, 2010-12-04, 2010-12-05, 20…
#> $ temp_c <dbl> 1.1638020, 1.3442852, 1.2533443, 1.0068728, 1.2899153, 1.229158…
glimpse(woody_island)
#> Rows: 735
#> Columns: 2
#> $ date <chr> "8/11/1990", "8/12/1990", "8/13/1990", "8/14/1990", "8/15/1990"…
#> $ temp_c <dbl> 25.850000, 23.308333, 18.533333, 15.350000, 13.966667, 11.35833…For your own data, assuming you have a .csv file in a
data folder in your working directory called
your_data.csv, you would call:
Using read.csv()
If you import your data in with functions like
read.csv() or read.table(), date columns will
be formatted as a characters (<chr>). You will have convert to
this column to a <date> or <dttm> type, and we recommend you
do this using lubridate, which makes dealing with date
a litter easier.
Below is an example of how you might do this. First, we check the
structure of the crooked_river and
woody_island datasets:
crooked_river <- read.csv(path_cr)
woody_island <- read.csv(path_wi)
glimpse(crooked_river) # note date column imported as a character (<chr>)
#> Rows: 1,826
#> Columns: 2
#> $ date <chr> "2010-12-01T00:00:00Z", "2010-12-02T00:00:00Z", "2010-12-03T00:…
#> $ temp_c <dbl> 1.1638020, 1.3442852, 1.2533443, 1.0068728, 1.2899153, 1.229158…
glimpse(woody_island) # note date column imported as a character (<chr>)
#> Rows: 735
#> Columns: 2
#> $ date <chr> "8/11/1990", "8/12/1990", "8/13/1990", "8/14/1990", "8/15/1990"…
#> $ temp_c <dbl> 25.850000, 23.308333, 18.533333, 15.350000, 13.966667, 11.35833…Then, we convert the date columns using lubridate:
# if your date is in the form "2000-09-01 12:00:00"
crooked_river$date <- ymd_hms(crooked_river$date)
# if your date is in the form "2000-09-01"
woody_island$date <- mdy(woody_island$date)
glimpse(crooked_river)
#> Rows: 1,826
#> Columns: 2
#> $ date <dttm> 2010-12-01, 2010-12-02, 2010-12-03, 2010-12-04, 2010-12-05, 20…
#> $ temp_c <dbl> 1.1638020, 1.3442852, 1.2533443, 1.0068728, 1.2899153, 1.229158…
glimpse(woody_island)
#> Rows: 735
#> Columns: 2
#> $ date <date> 1990-08-11, 1990-08-12, 1990-08-13, 1990-08-14, 1990-08-15, 19…
#> $ temp_c <dbl> 25.850000, 23.308333, 18.533333, 15.350000, 13.966667, 11.35833…Temperature data checks
After data are loaded and dates are formatted correctly, we can complete a series of checks to ensure that the temperature data is in the correct format and that there are no missing values.
hatchR provides several helper functions to assist with this process.
Visualize your temperature data
hatchR comes with the function
plot_check_temp() to visualize your imported data and
verify nothing strange happened during your import process. The function
outputs a ggplot2 object which can be subsequently
modified by the user. The arguments temp_min = and
temp_max = can be used to custom set thresholds for
expected temperature ranges (defaults are set at 0 and 25 °C). Here is
an example using the built-in dataset crooked_river:
plot_check_temp(data = crooked_river,
dates = date,
temperature = temp_c,
temp_min = 0,
temp_max = 12)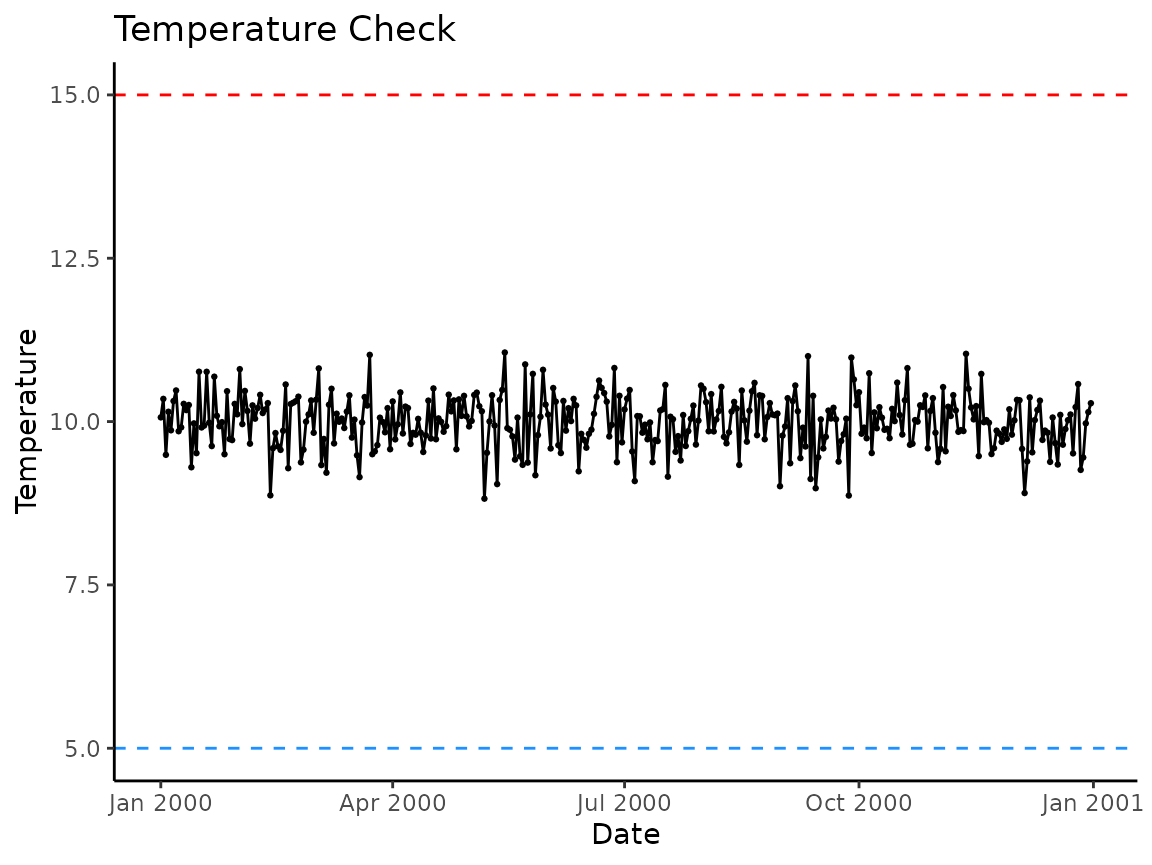
Summarize temperature data
If you imported raw data with multiple recordings per day,
hatchR has a built in function to summarize those data
to a daily average mean called summarize_temp(). The output
of the function is a tibble with mean daily temperature and its
corresponding.
Below, we simulate a dataset with 30-minute temperature recordings for a year and summarize the data to daily means.
# set seed for reproducibility
set.seed(123)
# create vector of date-times for a year at 30 minute intervals
dates <- seq(
from = ymd_hms("2000-01-01 00:00:00"),
to = ymd_hms("2000-12-31 23:59:59"),
by = "30 min"
)
# simulate temperature data
fake_data <- tibble(
date = dates,
temp = rnorm(n = length(dates), mean = 10, sd = 3) |> abs()
)
# check it
glimpse(fake_data)
#> Rows: 17,568
#> Columns: 2
#> $ date <dttm> 2000-01-01 00:00:00, 2000-01-01 00:30:00, 2000-01-01 01:00:00, 2…
#> $ temp <dbl> 8.318573, 9.309468, 14.676125, 10.211525, 10.387863, 15.145195, 1…Now we can summarize the data to daily means using
summarize_temp():
fake_data_sum <- summarize_temp(data = fake_data,
temperature = temp,
dates = date)
nrow(fake_data) #17568 records
#> [1] 17568
nrow(fake_data_sum) #366 records; 2000 was a leap year :)
#> [1] 366We again recommend, at a minimum, visually checking your data once it has been summarized.
# note we use fake_data_sum instead of fake_data
plot_check_temp(data = fake_data_sum,
dates = date,
temperature = daily_temp,
temp_min = 5,
temp_max = 15)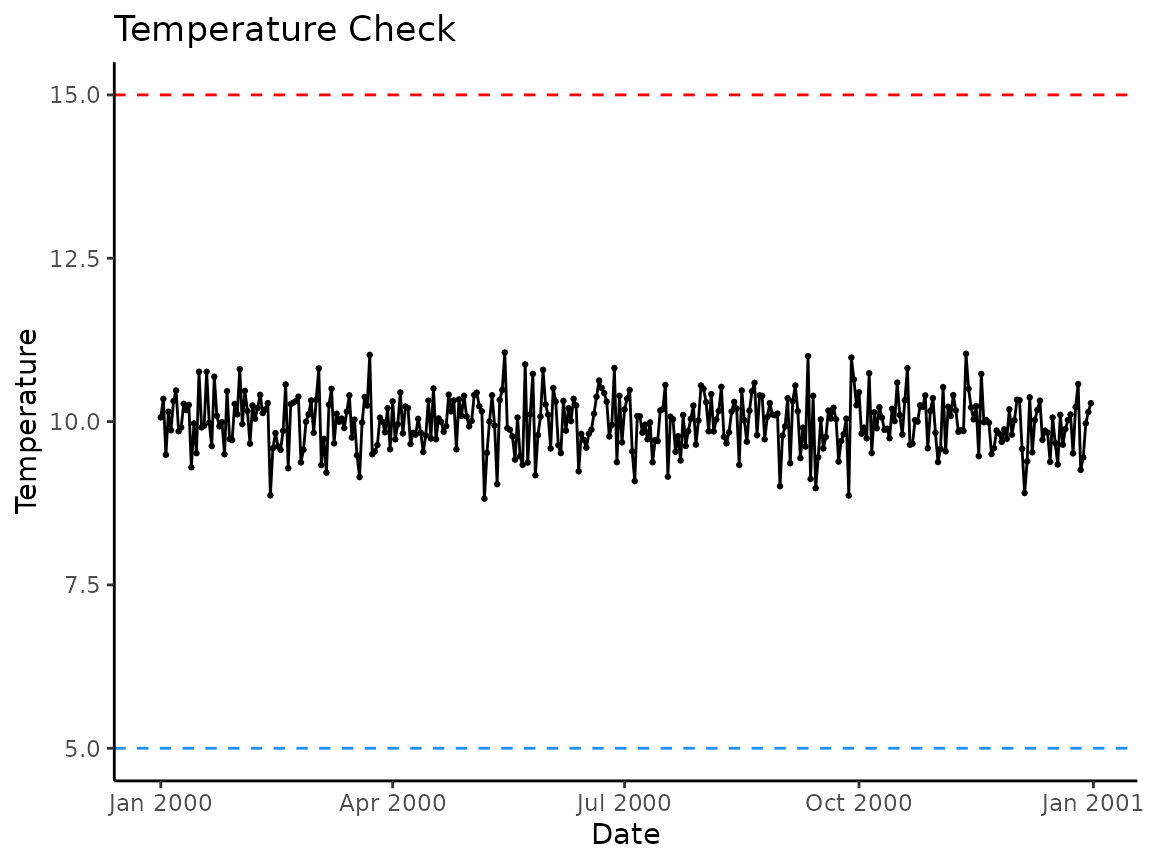
Check for continuous data
At present, hatchR only uses continuous data. Therefore, your data is expected to be continuous and complete.
You can check whether your data is complete and continuous using the
check_continuous() function. This function will return a
message if the data is or is not continuous or complete. Below, the
calls return all clear:
check_continuous(data = crooked_river, dates = date)
#> ℹ No breaks were found. All clear!
check_continuous(data = woody_island, dates = date)
#> ℹ No breaks were found. All clear!But if you had breaks in your data:
check_continuous(data = crooked_river[-5,], dates = date)
#> Warning: ! Data not continuous
#> ℹ Breaks found at rows:
#> ℹ 5You’ll get a note providing the row index where breaks are found for inspection.
If you have days of missing data, you could impute them using rolling means or other approaches.
Species-specific model parameters
hatchR has two options for selecting parameterized models for predicting fish early life history phenology using:
- model parameterizations included in the package
- custom parameterizations using your own data
This processes is described in the vignette Parameterize hatchR models
Spawn Dates
A date or range of dates for spawning the species of interest is required to predict phenology. This information can be provided in the form of a single date or a range of dates. More information on how to provide this information is available in the vignette Predict phenology: basic, as well as subsequent vignettes.
Next steps
After importing, wrangling, and checking your temperature data, there are a number of different actions you can take. These steps are laid out in subsequent vignettes: